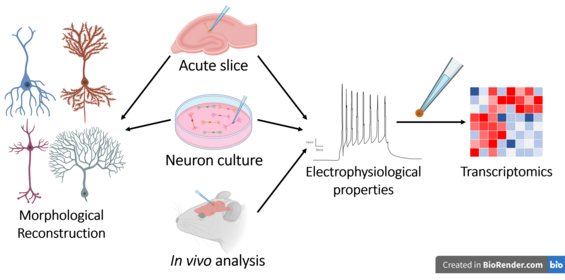
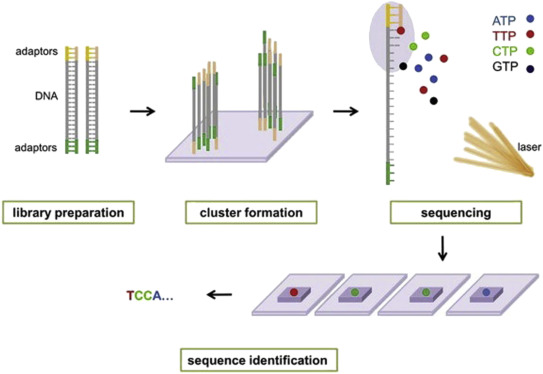
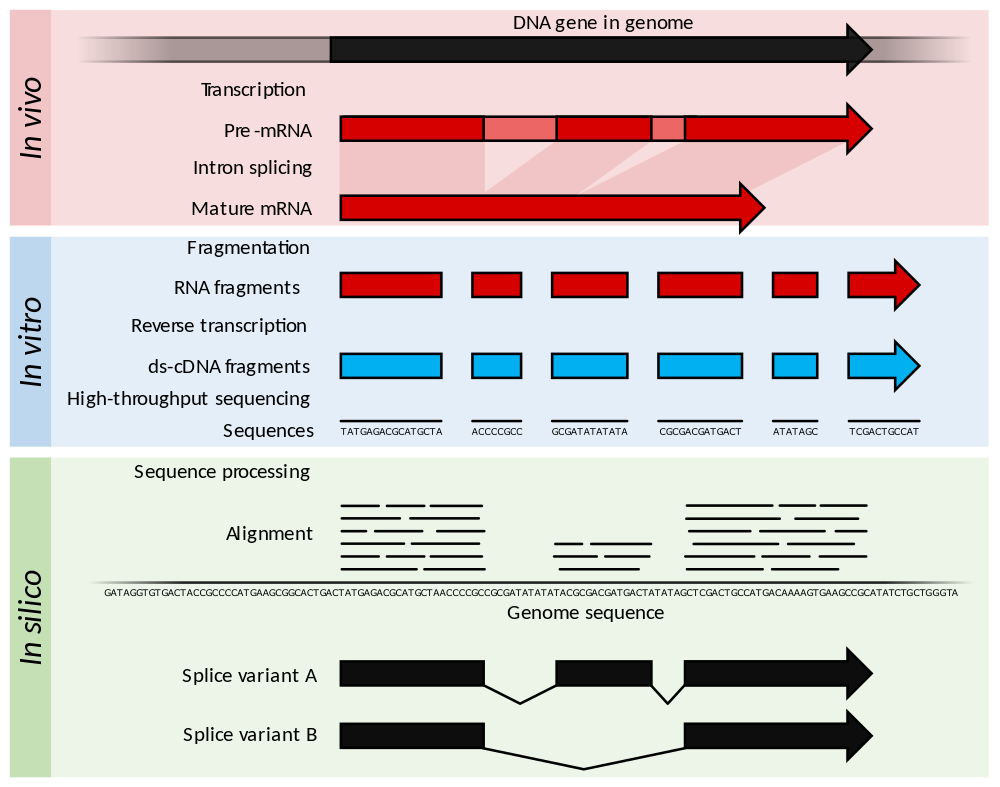
paired end sequencing wikipedia
For mRNA-Seq library prep use. The preparation of mate pair libraries is designed to allow classical paired-end sequencing of both ends of a fragment with an original size of several kilobases.

What Are Paired End Reads The Sequencing Center
No unintended insertion or backbone sequence of the transformed plasmid is observed at the whole genome level.
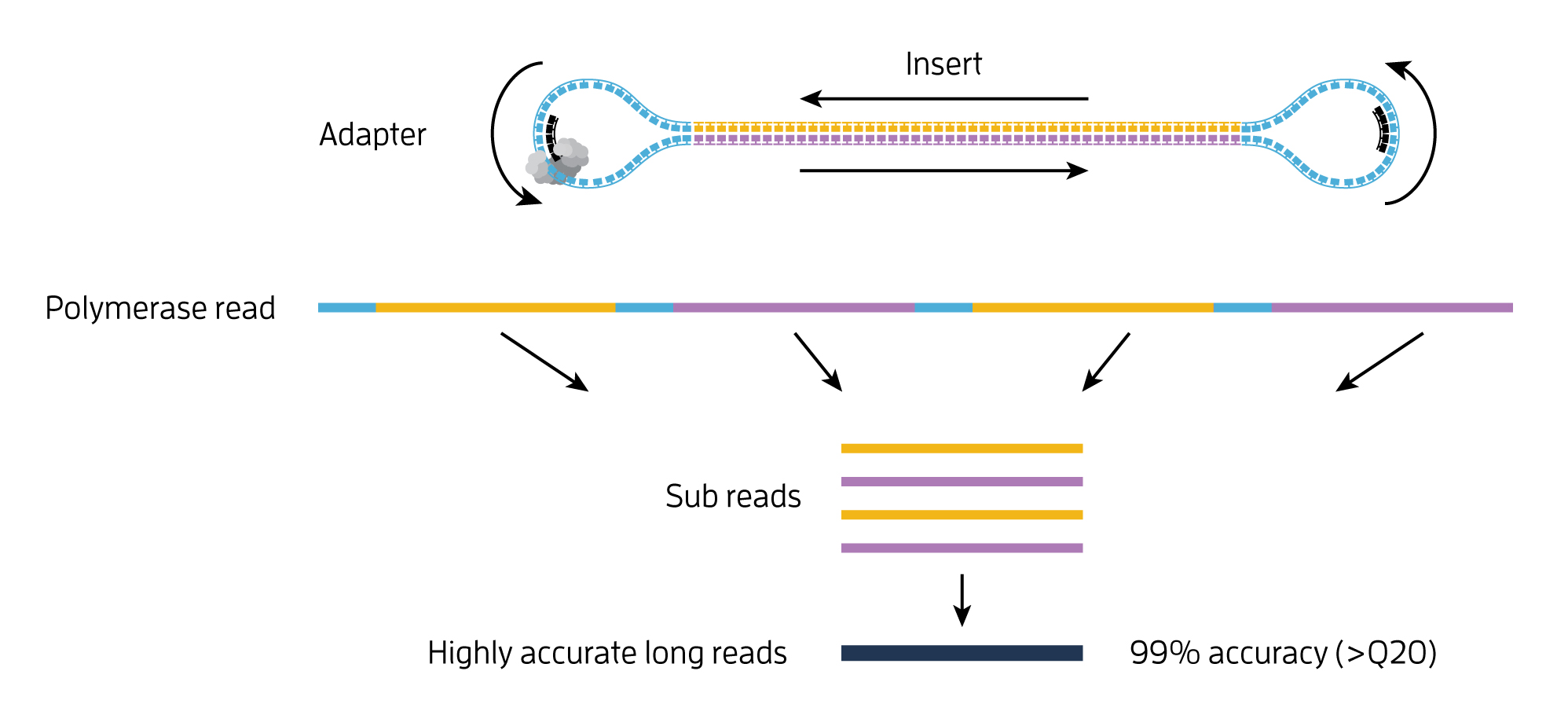
. Paired-end RNA sequencing RNA-Seq enables discovery applications such as detecting gene fusions in cancer and characterizing novel splice isoforms. Paired-end RNA sequencing RNA-Seq enables discovery applications such as detecting gene fusions in cancer and characterizing novel splice isoforms. Learn about the difference between Paired-End and Single-Run sequencing and why the former creates more precise alignments than the latter especiall.
Individual reads can be paired together to create paired-end reads which offers some benefits for downstream bioinformatics data analysis algorithms. Paired-end sequencing facilitates detection of genomic rearrangements and repetitive sequence elements as well as gene fusions and novel transcripts. Molecular characterization of the G281 event will.
The figure shows the workflow for mate-pair library preparation for Illumina sequencing. For paired-end RNA-Seq use the following kits with an alternate fragmentation protocol followed by standard Illumina paired-end cluster generation and sequencing. The structure of a paired-end read is described here.
T-DNA in a head-to-head arrangement. For example you shear up some genomic DNA and cut a region out at 500bp. Both are methodologies that in addition to the sequence information give you information about the physical distance between the two reads in your genome.
In short-read sequencing intact genomic DNA is sheared into several million short DNA fragments called reads. Combining data from mate pair sequencing with those from short-insert paired-end reads provides increased information for maximising sequencing coverage across a genome 1. A paired-end whole-genome sequencing approach enables comprehensive characterization of transgene integration in rice Commun Biol.
Mate pair sequencing is used for various applications applications including. This can be very helpful e. For paired-end RNA-Seq use the following kits with an alternate fragmentation protocol followed by standard Illumina paired-end cluster generation and sequencing.
For mRNA-Seq library prep use. However in many cases eg with Illumina NextSeq and NovaSeq. For your De novo genome assembly Fig.
Bases 1-75 forward direction and bases 225-300 reverse direction of the fragment. 2022 Jul 5. For example if you have a 300bp contiguous fragment the machine will sequence eg.
For the first test I took some sequence from the human genome hg19 and. Biocc paired end or mate pair refers to how the library is made and then how it is sequenced. Paired-end sequencing allows users to sequence both ends of a fragment and generate high-quality alignable sequence data.
The preparation of mate pair libraries is designed to allow classical paired-end sequencing of both ends of a fragment with an original size of several kilobases. In paired-end sequencing the library preparation yields a set of fragments and the machine sequences each fragment from both ends. Since the beginning of 2013 this preparation has been based on Nextera.
Since paired-end reads are more likely to align to a reference the. Typical experimental design advice for expression analyses using RNA-seq generally assumes that single-end reads provide robust gene-level expression estimates in a cost-effective manner and that the additional benefits obtained from paired-end sequencing are not worth the additional cost.
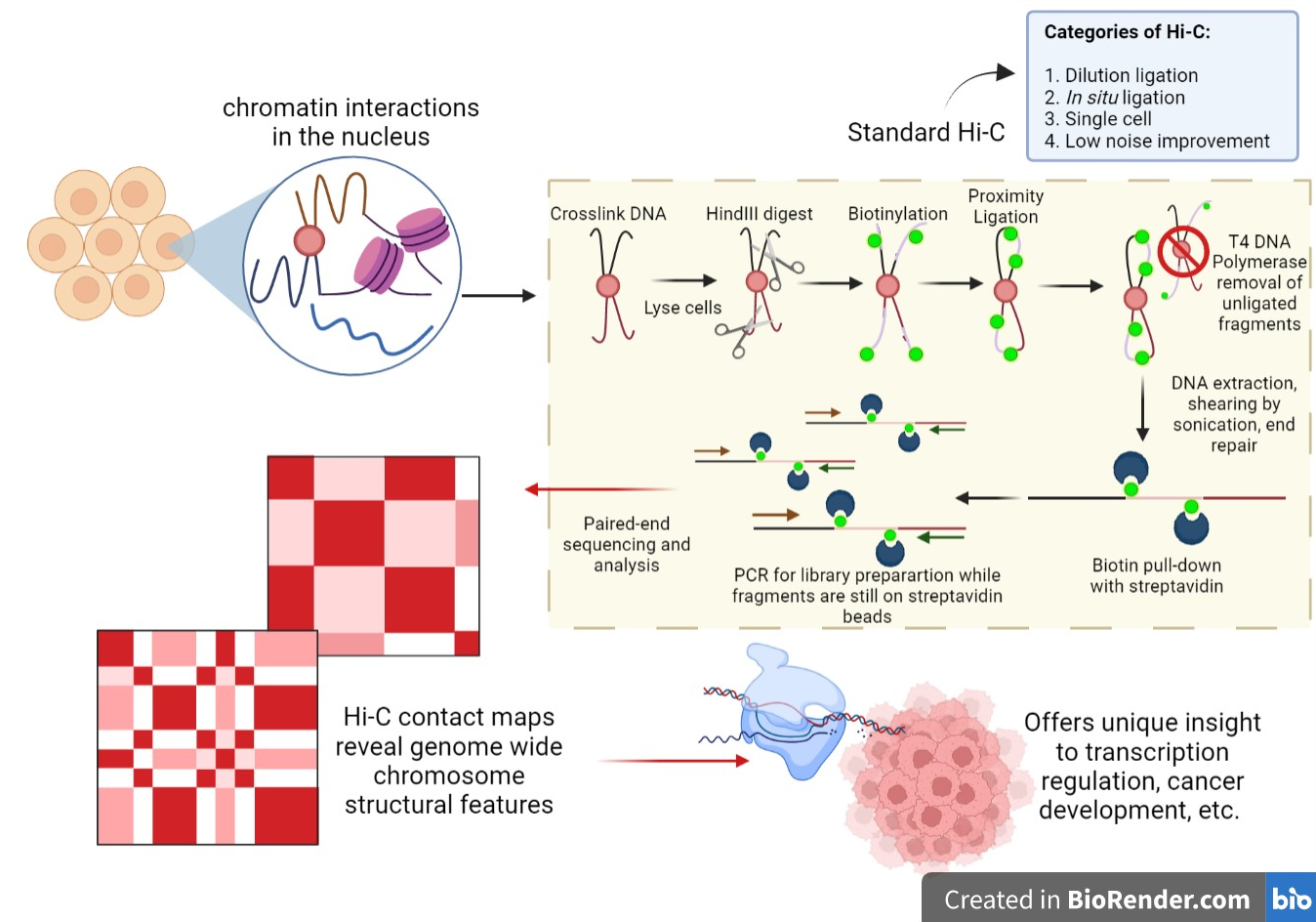
Hi C Genomic Analysis Technique Wikipedia

What Is Mate Pair Sequencing For
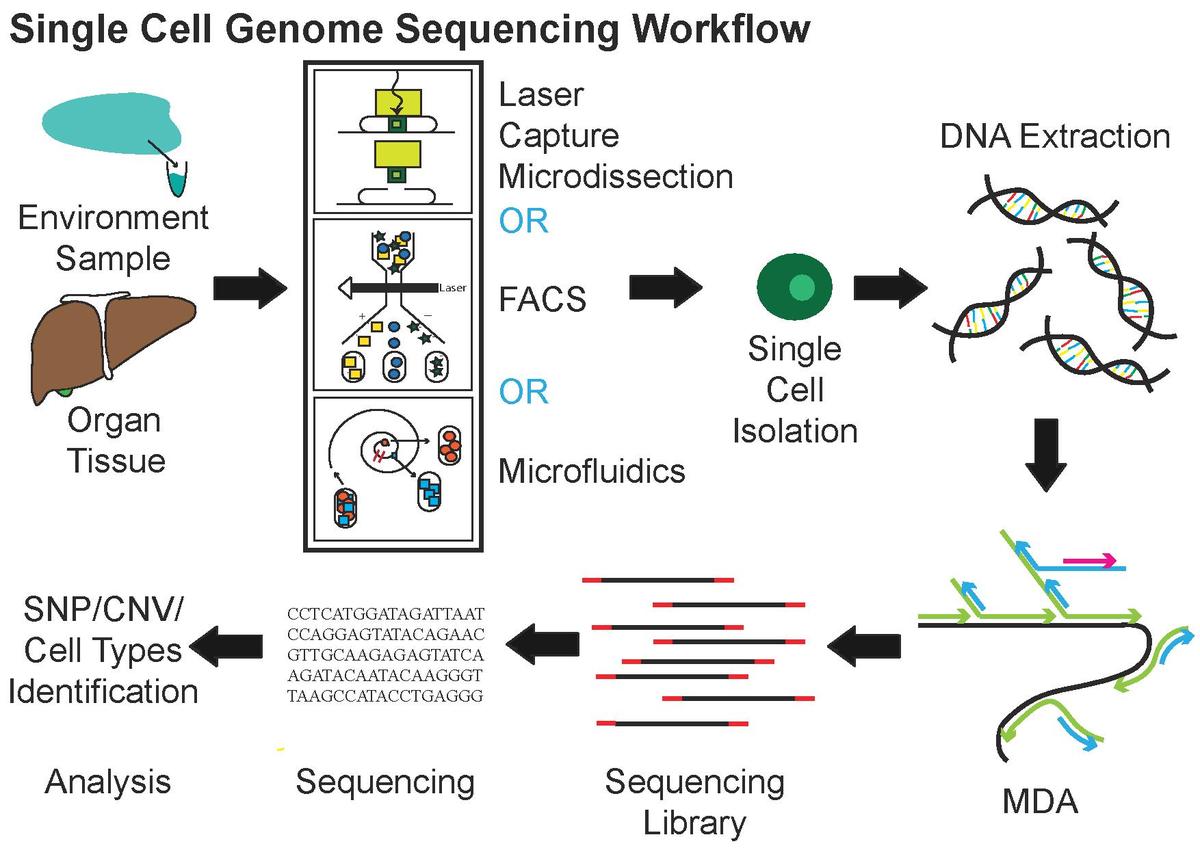
Chapter 1 Introduction To Single Cell Technology Fundamentals Of Scrnaseq Analysis

What Is Mate Pair Sequencing For
Difference Between Whole Genome Sequencing And Next Generation Sequencing Difference Between
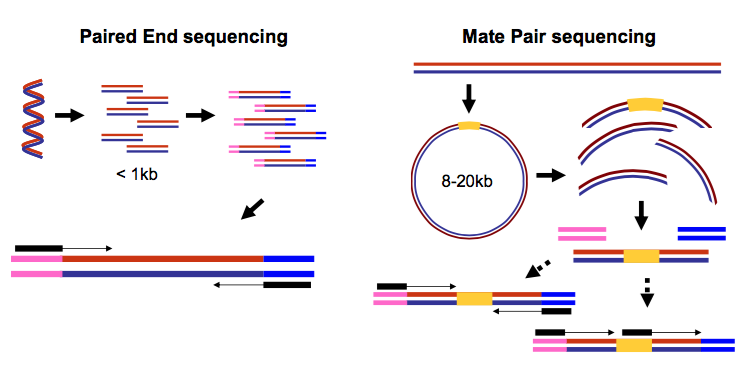
Manipulating Ngs Data With Galaxy Galaxy Community Hub
Proximal End Bias From In Vitro Reconstituted Nucleosomes And The Result On Downstream Data Analysis Plos One
Difference Between Whole Genome Sequencing And Next Generation Sequencing Difference Between